QCG Example 3
An example for constrained QCG calculations.
Constraining the solute
Sometimes GFN2-xTB or GFN-FF geometry optimizations might distort DFT optimized structures. To prevent this, it is possible to constrain the solute geometry by adding a file .xcontrol with constraints to the calculation. A typical example where this is necessary are transition metal molecules, such as the iron complex below.
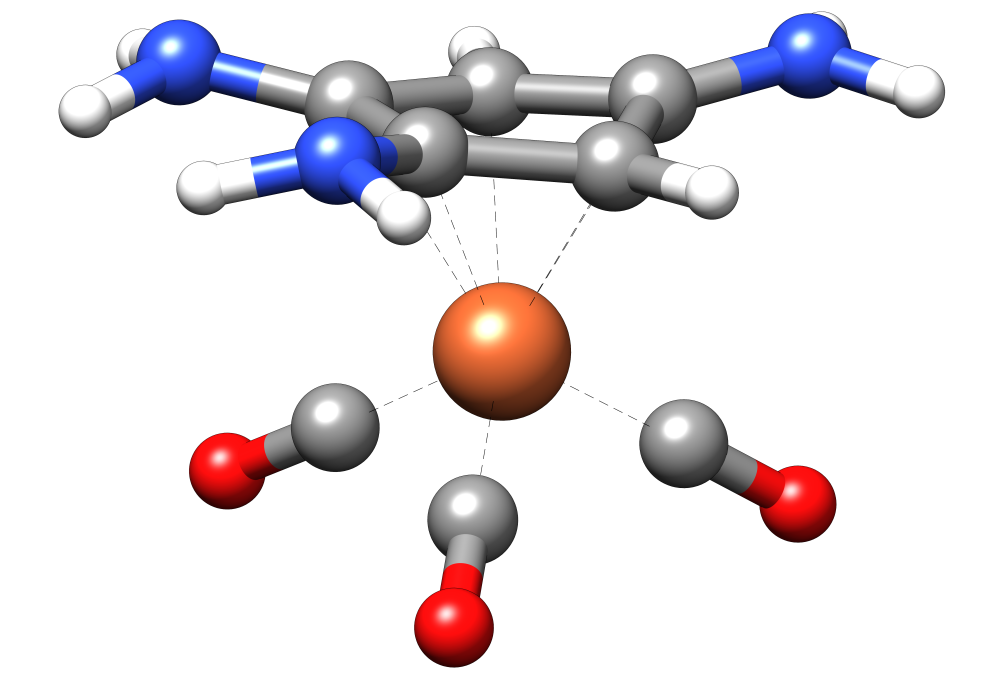
.xcontrol. The file is read automatically by the QCG procedure and can’t be specified separately via the command line.Now we want to constrain the ligands and to generate an ensemble. To do so, we provide the following .xcontrol file that constrains all bonds between the iron (16), the carbon atoms of the ring (atoms 3,4,6,7,8), and the CO ligands (atoms 17-22):
$constrain
atoms: 3,4,6-8,16,17-22
$end
Having prepared a directory with this file named .xcontrol, the solute coordinates solute.xyz, and the solvent coordinates solvent.xyz (acetonitrile), the ensemble is now generated for example with
crest solute.xyz --qcg solvent.xyz --nsolv 6 --T 12 --ensemble --gbsa h2o --mdtime 50 --mddump 200
23
N 1.3802608000 -0.0318528000 0.0463356000
N -0.4099459000 -2.4279732000 -0.4426793000
C -0.8233287000 -1.1730691000 -0.1562094000
C 0.0282237000 -0.0329935000 0.0761397000
N -3.1128965000 1.1237167000 1.4357707000
C -0.8148438000 1.0531548000 0.5128170000
C -2.1758731000 -0.7690355000 0.1411386000
C -2.1257237000 0.5229799000 0.7675646000
H -0.4572325000 2.0269118000 0.8223111000
H -4.0464379000 0.7439366000 1.3737637000
H -3.0303702000 2.1068597000 1.6506659000
H -1.1169391000 -3.0848452000 -0.7424315000
H 0.4822106000 -2.5429436000 -0.9026828000
H 1.8456312000 -0.7155480000 -0.5345244000
H 1.8360609000 0.8698550000 0.0667192000
H -3.0359762000 -1.4262449000 0.1182750000
Fe -1.5405186000 0.5270943000 -1.3978036000
C -1.8580173000 -0.6232521000 -2.7083661000
C -2.9303295000 1.6215990000 -1.6740517000
C -0.3124958000 1.4435812000 -2.2886848000
O 0.5082387000 2.0393586000 -2.8321404000
O -2.0651590000 -1.4011695000 -3.5304385000
O -3.8189784000 2.3247399000 -1.8686288000
6
C -5.1936370000 1.8144547000 -0.0000255000
C -3.7637653000 1.6301290000 -0.0000193000
H -5.5336302000 2.0849167000 0.9967301000
H -5.6871686000 0.8944148000 -0.3037251000
H -5.4665453000 2.6066235000 -0.6930206000
N -2.6155337000 1.4820613000 0.0000603000
Two ensembles will result:
- a “complete” one named
full_ensemble.xyz - and one with only the highest populated clusters
final_ensemble.xyz)
All ensembles will have a solute structure that has the same Fe-C geometries as the solute.xyz file.
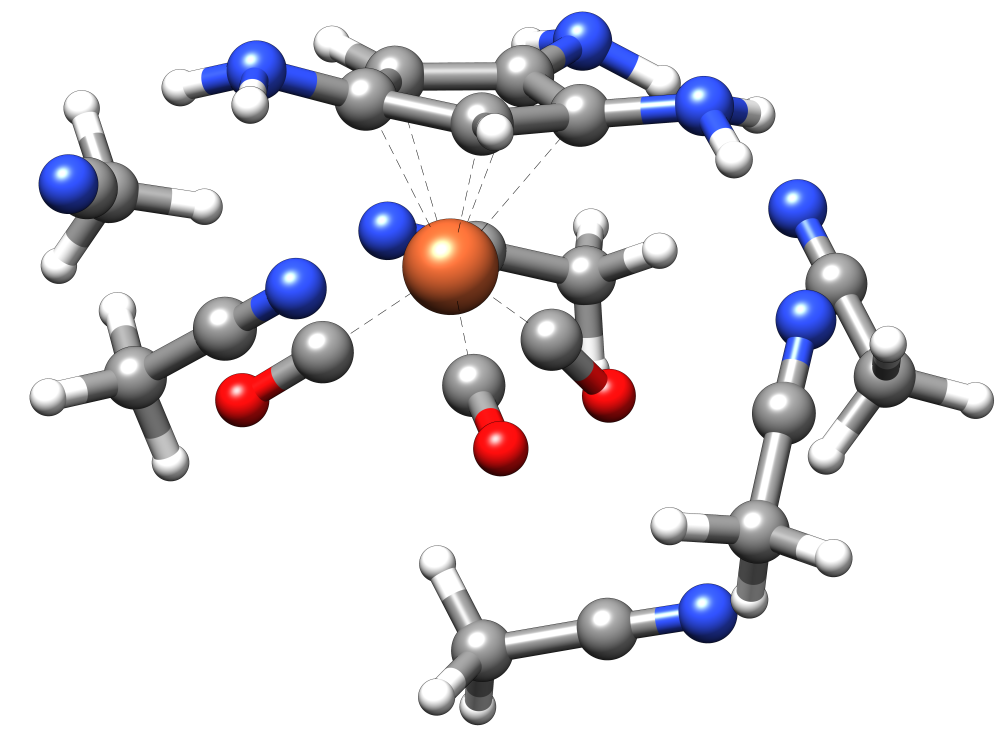
--nopreopt.