TS Conformers
An example for sampling conformers at the transition-state.
Sampling of conformations at the transition-state
At first, a transition-state (TS) has to be localized. Then, the TS mode has to be identified and reasonable constraints have to be applied to freeze this mode during the CREST run. Choosing suitable constraints is the responsibility of the user! For a detailed guide on how to add constraints see Example 4.
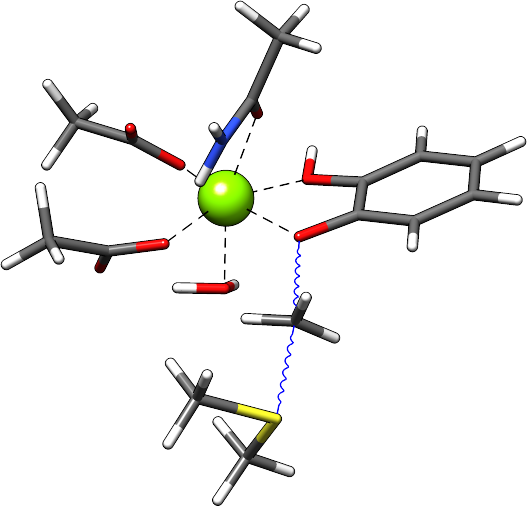
In this example, a methyl group is transferred onto the catechol molecule. To preserve the TS vibrational mode the atoms which are dominantly contributing to this mode are fixed. In this case, the carbon (atom #36) of the methyl group being transferred, the sulfur (atom #37) of the S-adenosyl-L-methionine (SAM) and the oxygen (atom #35) of the catechol group are constrained. For running the TS conformational search only these atoms have to be constrained. But to retain the surrounding enzyme environment additionally the distances of all ligands to the magnesium cation and the amide magnesium water angle were constrained. As stated before all atoms with constraints have to be removed from the list of atoms which are used in the metadynamics simulation.
crest coord --cinp constraints.inp --gbsa methanol
$coord
-2.57480197685137 -0.38573933229522 0.86228536590435 Mg
-5.87996595426622 -1.46598597135567 -1.00931632324148 O
-5.79755045954234 1.88737481602186 1.36486580018227 O
-6.93504356011937 0.41703174067196 -0.07677235660280 C
-9.68583177367761 0.93957235453071 -0.70260934507636 C
-9.88785370898918 2.90051382662291 -1.27585066001173 H
-10.31204304615949 -0.31693795001232 -2.19707799857187 H
-10.81224558069477 0.63532604630470 0.98871505743889 H
-1.35732893615725 2.84149984259631 3.74273757259152 O
-1.31788637685368 1.88478932440519 -1.80336662588251 O
-1.03506712269361 -3.09136305475668 -1.65209468828016 O
-3.01034174150676 3.35231258504990 4.30691490291278 H
-0.64007292100150 4.31049584542225 2.93186531615926 H
-3.02042382593105 -2.69109360436689 3.78441246580865 O
-0.67413309122153 -2.78784634989936 4.10013037720282 C
0.80704125300360 -1.59087682326574 2.72475235410942 O
0.37030033373577 -4.45667671167827 6.17913372417457 C
1.65729077111170 -3.36053569450090 7.34278701173010 H
-1.17079464125707 -5.18933342363882 7.31676317209597 H
1.41212360996512 -6.00880794547076 5.32805483610633 H
-0.04610218809699 0.99217247488345 -2.84947633284740 H
-0.58166801572397 4.35407649708453 -2.13719082516246 C
1.69930763718877 4.60968100984284 -3.53188509022323 C
-1.89895861199073 6.41295502711680 -1.26089925937752 C
2.61815567802848 7.04758861150735 -3.94211016089909 C
-0.94293511850593 8.82264113991643 -1.71734825726509 C
-3.65794447068903 6.13213826999732 -0.25859371242962 H
1.29133066638906 9.11831895867148 -3.04019344765619 C
4.35136261809131 7.29515670682662 -4.99253235854911 H
-1.96139641783255 10.45433175989920 -1.03894063047482 H
2.01793975704253 10.99527109251927 -3.38477251662235 H
5.63677744964081 -0.19526366812337 -3.54734464996746 H
3.55857435122244 0.44545364581733 -0.79647639427433 H
6.02794370271953 2.75567866080431 -1.74563412676399 H
2.74773927853638 2.50310064429053 -4.32763740793204 O
5.16232303152189 0.93488296527549 -1.93713143185301 C
7.77908129622702 -0.95480533027442 0.60724611364076 S
6.20470140355368 -3.99408071134196 0.68137239550646 C
7.00770708640275 -5.10883646299712 2.20213746286551 H
4.19551348270129 -3.68373090740626 0.97362752914345 H
6.54643468112530 -4.90904155689111 -1.11917138292065 H
6.61325357496481 0.34737209228094 3.55003016825311 C
7.52593267335208 -0.62757026577676 5.10500275305939 H
7.10342021330197 2.33658535430792 3.58672294810726 H
4.57513571292400 0.10172782556556 3.62256009227771 H
-1.61022171124489 -5.31411191371024 -2.02789529853598 C
-3.17527947979499 -6.57718946281529 -0.51674594958634 N
-3.77763814894346 -8.33207207055257 -0.93763600526181 H
-4.05833804986482 -5.57635320116590 0.85099090510650 H
-0.47266612030322 -6.78426594278943 -4.18601622917577 C
0.51805850799787 -8.43374379675092 -3.46937160488911 H
-1.96305386150678 -7.41025810365247 -5.45278966275112 H
0.83013814067146 -5.58152886274452 -5.21822759129119 H
$end
$constrain
atoms: 35-37
force constant=0.5
reference=coord.ref
distance: 10, 1, auto
distance: 2, 1, auto
distance: 11, 1, auto
distance: 14, 1, auto
distance: 9, 1, auto
angle: 9, 1, 11, 180
$metadyn
atoms: 3-8,12-13,15-34,38-53
$end
The TS conformer search yields 141 conformers within 6 kcal/mol. For each of these conformers, Hessians have to be calculated to ensure that the transition-state mode is preserved. Those conformers with preserved mode can be optimized into the TS and the true TSs have to be confirmed by again a Hessian calculation (only one imaginary mode). During the optimization, some conformers can become identical or rotamers of each other. To this end, all optimized geometries are appended and sorted with the cregen sorting routine.
cat TSconf*.xyz >> allts.xyz
crest coord -cregen allts.xyz -ewin 30
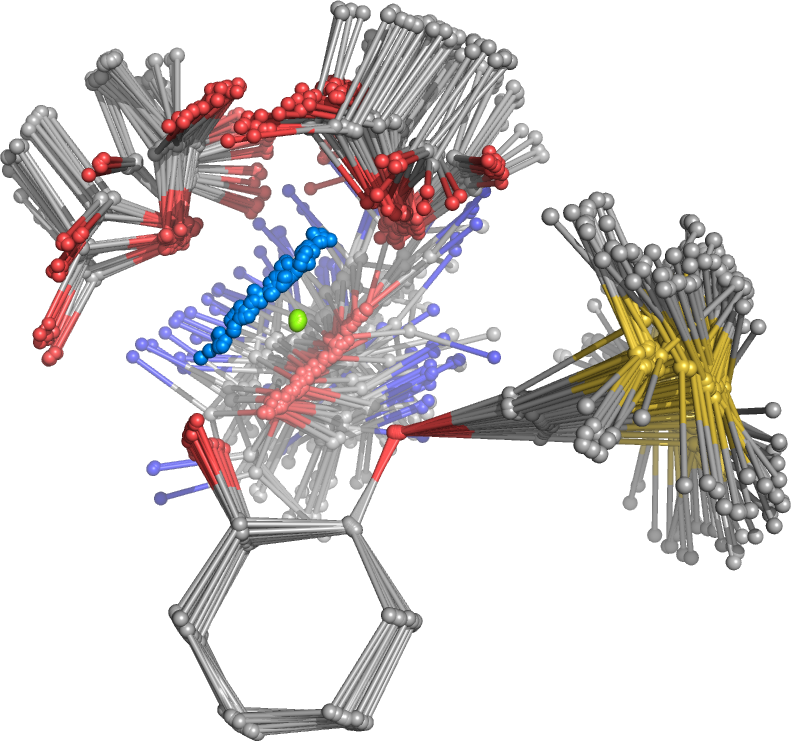
After sorting only 91 unique TS conformers are obtained within an energy window of 6.1 kcal/mol. This procedure can in principle be refined at DFT level.