QCG Example 1
An example for building up solvent shells around a molecule.
Building up solvent shells for an MD simulation
Explicitly modeled solvent molecules can lead to different properties of the solute compared to implicit solvent models. Thus, it might be necessary to check on this, for example, for geometries and IR spectra.
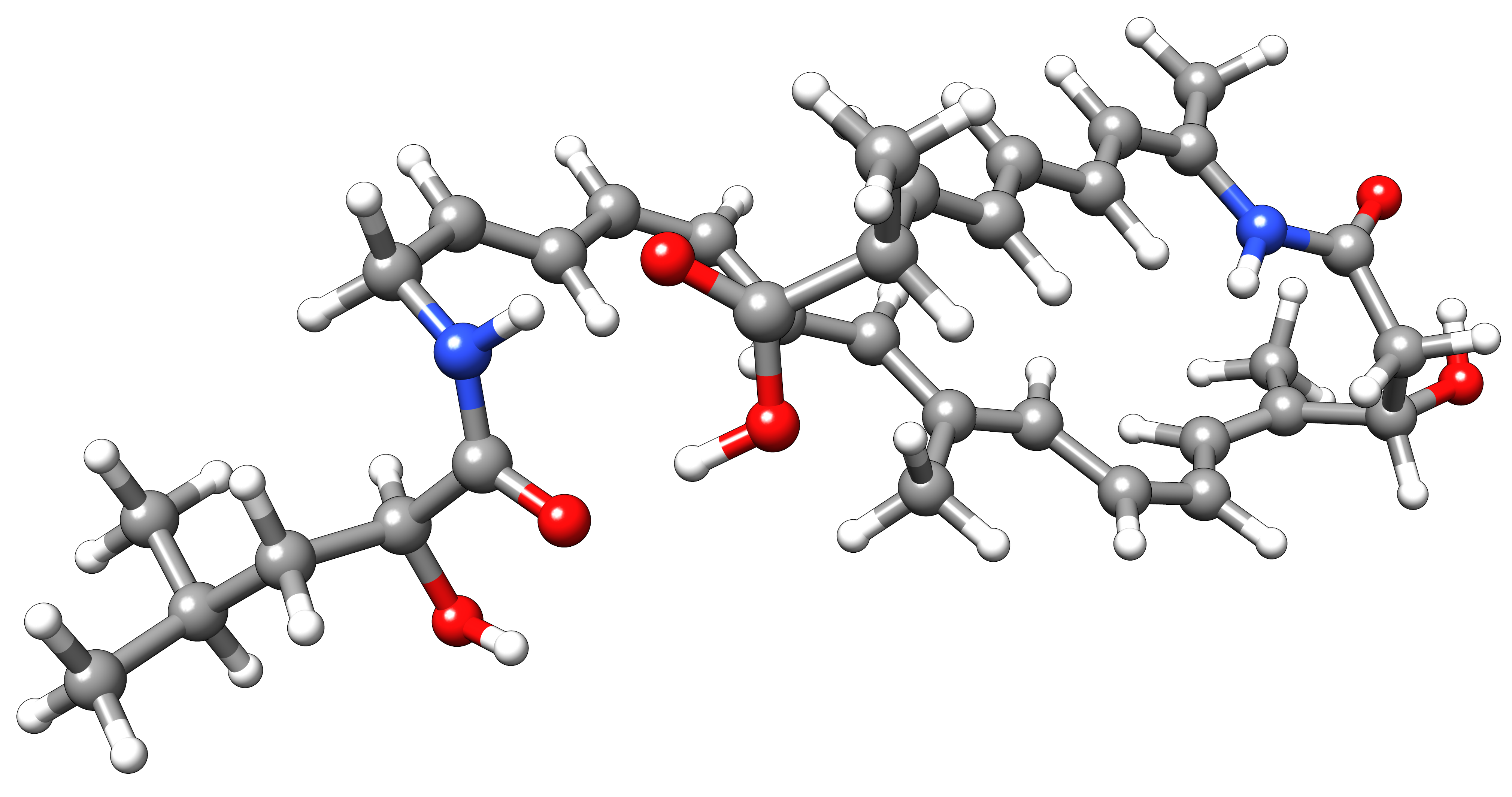
Let’s assume we have the bacillaene molecule and want to grow a cluster of 100 water molecules around it. Afterward, we perform an MD simulation to investigate the geometry in solution. To do so, we provide input coordinates of the solute bacillaene.xyz and of a water molecule water.xyz. As we do not have much time, GFN-FF is also used during the growth algorithm. We call CREST and activate the QCG algorithm with the following command.
crest bacillaene.xyz --qcg water.xyz --nsolv 100 --gfnff --T 12 --alpb water --nofix
90
C -5.3127996594 -2.4157946011 -0.5090291244
C -6.6369198591 -2.2744765141 -0.3505867691
C -4.5376337067 -1.9989947690 -1.6511538708
C -7.4911082799 -1.4906802100 -1.3353795445
C -7.3417027536 -2.8130153570 0.8534300295
C -6.9820968905 -0.0260037238 -1.4231824713
C -3.2303223550 -1.6498120479 -1.6254976982
C -2.4648530707 -1.4782784301 -0.4345685359
C -1.1490119425 -1.1466912716 -0.3267799699
C -0.2345729980 -0.9625082034 -1.4997955626
C -0.6049768456 -0.9722141905 0.9890332154
C 0.6701604636 -0.6330961686 1.2870390935
C 1.1341096011 -0.4761647542 2.6269132552
C 2.3830259753 -0.1235992724 3.0110310022
C 3.4734843635 0.1679005598 2.1268262297
C 4.6931387856 0.5393278145 2.5267510375
C 5.8304304131 0.8677548326 1.6003866159
N 5.4283570547 0.8855111783 0.2104033711
C 5.3166273681 -0.1533114661 -0.6067536733
C 6.0134079054 -1.4661798013 -0.2726105716
O 4.7074640990 -0.0705419639 -1.6861449989
C 7.5144666461 -1.3152943375 -0.5941160582
O 5.4313708211 -2.5035661443 -1.0272412069
C -6.7935147347 0.4641208344 -0.0003944175
O -7.7403733383 0.5228053447 0.7742620645
N -5.5106869286 0.7636403711 0.3110847277
C -4.9463300150 1.1535464336 1.5274406520
C -3.6739854047 1.6059179412 1.5754484283
C -5.7878061510 1.0225637475 2.7522984886
C -2.7844664843 1.8243840709 0.4767015254
C -1.5364316121 2.3145689941 0.6208013650
C -0.6655220004 2.5888732490 -0.4843830298
C 0.5487325406 3.1343609346 -0.3631391694
C 8.3298335983 -2.5431938358 -0.1704211284
C 9.6891088069 -2.5276652589 -0.8678074623
C 8.5197655075 -2.5906603796 1.3443209467
O -8.8586444577 -1.4991160896 -1.0023350180
C 1.4399210852 3.4740558350 -1.5251049648
C 2.6756050844 2.6104389356 -1.3716483915
C 1.8111558123 4.9585806785 -1.5160495647
O 2.6206023923 1.5056225590 -2.0814961045
O 3.6041031244 2.8850885925 -0.6340527724
H -4.7625204190 -2.9056795986 0.2836333196
H -5.0509303359 -2.0042601899 -2.6049219243
H -7.4349047919 -1.9513459821 -2.3302603369
H -6.6760991929 -3.4329009729 1.4477280383
H -8.2048607459 -3.4019112683 0.5445011777
H -7.7116749441 -1.9955343308 1.4750390520
H -6.0619084015 0.0254594956 -1.9985287387
H -7.7560134130 0.5683074568 -1.9132455671
H -2.7489218428 -1.4394091975 -2.5706796840
H -3.0129717265 -1.5960544885 0.4909743144
H -0.7561184183 -1.0725573851 -2.4442035526
H 0.5638309381 -1.7039098370 -1.4666980415
H 0.2258577807 0.0242190429 -1.4708519245
H -1.2956296326 -1.1246557687 1.8098949728
H 1.3745898015 -0.4696821456 0.4862918222
H 0.3981454779 -0.6600516684 3.4004310551
H 2.5974468939 -0.0405121044 4.0685973172
H 3.2726215465 0.0815655312 1.0682418239
H 4.9262739703 0.6324234323 3.5775689496
H 6.6468639108 0.1562857796 1.7478921113
H 6.2119507967 1.8653636049 1.8407595221
H 4.8859889259 1.7105149385 -0.0683642472
H 5.8575220450 -1.7269366655 0.7774134480
H 7.6007536957 -1.1797242272 -1.6752148153
H 7.9082834268 -0.4205120198 -0.1104506371
H 5.0955504465 -2.0978322698 -1.8442489323
H -4.8389468734 0.6790182836 -0.4409166158
H -3.2785159256 1.8587339956 2.5484209564
H -5.2246095910 1.3178627663 3.6320617997
H -6.1254267820 -0.0065165747 2.8692478768
H -6.6790660370 1.6429351108 2.6701403518
H -3.1292138183 1.6120944762 -0.5273979730
H -1.1579767117 2.5402684410 1.6084450304
H -1.0491040613 2.3573854617 -1.4703055793
H 0.9505481083 3.3742128511 0.6120025660
H 7.7785462919 -3.4349097892 -0.4874864977
H 9.5694109024 -2.5429149313 -1.9488155636
H 10.2730806739 -3.3975457026 -0.5768929878
H 10.2450015890 -1.6328073712 -0.5947376585
H 9.0991604017 -3.4685164813 1.6200641190
H 7.5644928769 -2.6459407744 1.8604244387
H 9.0545665098 -1.7068730142 1.6874964687
H -8.9584252351 -0.9401749581 -0.2142721535
H 0.9490038267 3.1991238510 -2.4619332618
H 2.4126870178 5.2036544364 -2.3883354398
H 2.3855713372 5.1906177538 -0.6223783473
H 0.9080079216 5.5625777336 -1.5263116425
H 3.4359643823 0.9211814126 -1.9665264904
3
O -0.1918040235 1.3862489483 0.0047370042
H 0.7660977787 1.3911615443 -0.0141642652
H -0.4927337474 1.6150799341 -0.8756928250
==============================================
| |
| C R E S T |
| |
| Conformer-Rotamer Ensemble Sampling Tool |
| based on the GFN methods |
| P.Pracht, S.Grimme |
| Universitaet Bonn, MCTC |
==============================================
Version 2.11, Mon 19. Apr 11:43:20 CEST 2021
Using the xTB program. Compatible with xTB version 6.4.0
Cite work conducted with this code as
P. Pracht, F. Bohle, S. Grimme, PCCP, 2020, 22, 7169-7192.
and S. Grimme, JCTC, 2019, 15, 2847-2862.
with help from:
C.Bannwarth, F.Bohle, S.Ehlert, S.Grimme,
C. Plett, P.Pracht, S. Spicher
This program is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE.
Command line input:
> crest bacillaene.xyz -qcg water.xyz -gfnff -nsolv 100 -T 20 -grow -alpb water -nofix
Solute-file: bacillaene.xyz
Solvent-file: water.xyz
-gfnff : Use of GFN-FF requested.
-T 20 (CPUs/Threads selected)
Use of GFN-FF for ensemble search requested.
Use of GFN-FF for frequency computation requested.
-mdtime 10 (MD length in ps)
--alpb water : implicit solvation
========================================
| ---------------- |
| Q C G |
| ---------------- |
| Quantum Cluster Growth |
| University of Bonn, MCTC |
========================================
S. Grimme, S. Spicher, C.Plett, unpublished.
=========================================
| quantum cluster growth: INPUT |
=========================================
QCG: Only Cluster Generation
input parameters
solute : bacillaene.xyz
charge : 0
uhf : 0
solvent : water.xyz
# of solvents to add : 100
# of cluster generated : 4
# of CPUs used : 20
Solvation model : water
xtb opt level : normal
System temperature [K] : 298.0
RRHO scaling factor : 0.75
Solute geometry
molecular radius (Bohr**1): 11.20
molecular area (Bohr**2): 2554.19
molecular volume (Bohr**3): 5887.65
Solvent geometry
molecular radius (Bohr**1): 3.88
molecular area (Bohr**2): 194.90
molecular volume (Bohr**3): 244.27
radius of solute : 18.06
radius of solvent : 6.25
=========================================
| Preoptimization |
=========================================
-------------------------
xTB Geometry Optimization
-------------------------
Geometry successfully optimized.
Generating LMOs for solute
Total Energy of solute: -127.4297277 Eh
-------------------------
xTB Geometry Optimization
-------------------------
Geometry successfully optimized.
Generating LMOs for solvent
Total energy of solvent: -5.0703134 Eh
________________________________________________________________________
__________________ Solute Cluster Generation _____________________
________________________________________________________________________
=========================================
| quantum cluster growth: GROW |
=========================================
Solute:
unit ellipsoid axis a,b,c : 0.428 0.289 0.283
Solvent:
unit ellipsoid axis a,b,c : 0.384 0.323 0.292
solvent anisotropy : 1.130
solute anisotropy : 1.197
roff inner wall : 6.997
solute max dist : 39.969
solvent max dist : 7.298
inner unit axis : 0.528 0.241 0.231
inner ellipsoid/Bohr: 36.893 16.810 16.173
outer ellipsoid/Bohr: 28.654 19.342 18.972
Size E /Eh De/kcal Detot/kcal Density Efix R av/act. Surface Opt
1 -132.538524 -24.15 -24.15 1.115 -13.744 0.0 0.0 6017.1 normal
2 -137.624631 -9.91 -34.06 1.117 -14.046 9.9 8.9 6188.1 normal
3 -142.708487 -8.50 -42.56 1.120 -14.343 9.4 8.8 6350.0 normal
4 -147.796548 -11.14 -53.69 1.119 -14.634 9.2 14.3 6536.7 normal
[...]
100 -636.352408 -7.47 ******* 1.198 -32.223 14.2 14.9 22281.9 normal
Final gfn2 optimization
Growth finished after 100 solvents added
Results can be found in grow directory
Energy list on file 'qcg_energy.dat'
Interaction energy on file 'qcg_conv.dat'
Growing process on 'qcg_grow.xyz'
Final geometry after grow in 'cluster.coord' and 'cluster.xyz'
Potentials and geometry written in 'cluster_cavity.coord' and 'twopot_cavity.coord'
-----------------
Wall Time Summary
-----------------
Grow wall time : 0h :34m :32s
--------------------
Overall wall time : 0h :34m :32s
CREST terminated normally.
QCG automatically detects water as a solvent. This will cause the outer wall potential to be scaled to smaller sizes. Additionally, the solute will be fixed during the growth (also only in case of water). Bacillaene has a variety of different conformations and we want to consider the response of the intramolecular geometry upon addition of water. Thus, the --nofix flag was provided.
--nofix. For other solvents, the fixing the solute geometry can be requested by using the keyword --fixsolute.Notice that we also choose the ALPB water model to get energies including an additional implicit solvation potential that is surrounding the cluster. The resulting cluster can be found in the grow directory and cluster.xyz file. Additionally, each addition of a solvent molecule is written to qcg_grow.xyz.
As we want to perform an MD simulation on this structure without dissociating the cluster, we also need the wall potentials found in the wall_potential file. This can directly be used as an input for xtb to perform the constrained MD simulation.